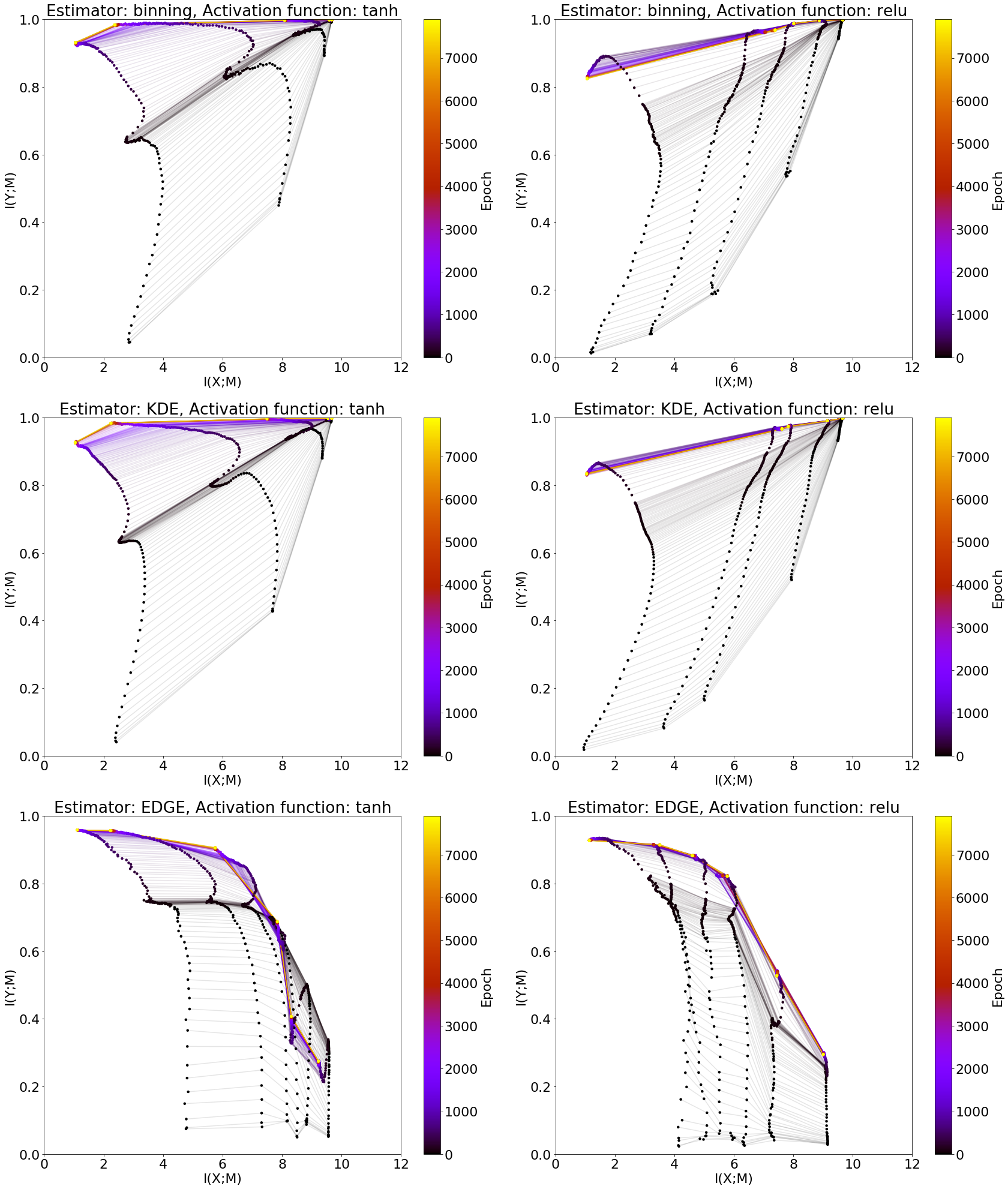
Comparison of infoplanes for different estimators¶
[1]:
import sys
sys.path.append('../..')
from deep_bottleneck.eval_tools.experiment_loader import ExperimentLoader
from deep_bottleneck.eval_tools.utils import format_config, find_differing_config_keys
import matplotlib.pyplot as plt
from io import BytesIO
[2]:
loader = ExperimentLoader()
[5]:
import numpy as np
import pandas as pd
all_experiments = [[1318,1328,1327,1320,1333,1315,1325,1319,1321,1323], # binning tanh
[1326,1329,1316,1330,1331,1317,1332,1334,1324,1322], # binning relu
[1368,1369,1370,1371,1372,1375,1373,1374,1376,1377], # kde tanh
[1354,1358,1359,1361,1362,1363,1364,1365,1367,1366], # kde relu
[1454,1441,1451,1453,1455,1448,1459,1442,1458,1447], # EDGE tanh
[1452,1445,1446,1440,1449,1443,1444,1457,1450,1456] # EDGE relu
]
label_list = ['Estimator: binning, Activation function: tanh',
'Estimator: binning, Activation function: relu',
'Estimator: KDE, Activation function: tanh',
'Estimator: KDE, Activation function: relu',
'Estimator: EDGE, Activation function: tanh',
'Estimator: EDGE, Activation function: relu',
]
all_means = []
for experiment_set in all_experiments:
experiments = loader.find_by_ids(experiment_set)
set_df = []
for i, experiment in enumerate(experiments):
infoplane = experiment.artifacts['information_measures_test'].show()
set_df.append(infoplane)
df_concat = pd.concat(set_df)
by_row_index = df_concat.groupby(df_concat.index)
set_mean = by_row_index.mean()
all_means.append(set_mean)
[6]:
plt.rcParams.update({'font.size': 22})
fig, ax = plt.subplots(figsize=(24,28))
for ax_id, set_mean in enumerate(all_means):
plt.subplot(3,2,ax_id+1)
measures = set_mean.groupby(['epoch', 'layer']).mean()
total_epochs = measures.index.get_level_values('epoch')[-1] + 1 # epoch index starts at 0
sm = plt.cm.ScalarMappable(cmap='gnuplot', norm=plt.Normalize(vmin=0, vmax=total_epochs))
sm.set_array([])
for epoch_nr, mi_measures in measures.groupby(level=0):
color = sm.to_rgba(epoch_nr)
xmvals = np.array(mi_measures['MI_XM'])
ymvals = np.array(mi_measures['MI_YM'])
plt.plot(xmvals, ymvals, color=color, alpha=0.1, zorder=1)
plt.scatter(xmvals, ymvals, s=20, facecolors=color, edgecolor='none', zorder=2)
plt.title(label_list[ax_id])
plt.xlabel('I(X;M)')
plt.ylabel('I(Y;M)')
plt.xlim([0, 12])
plt.ylim([0, 1])
fig.colorbar(sm, label='Epoch')
plt.tight_layout()
plt.show()
[ ]:
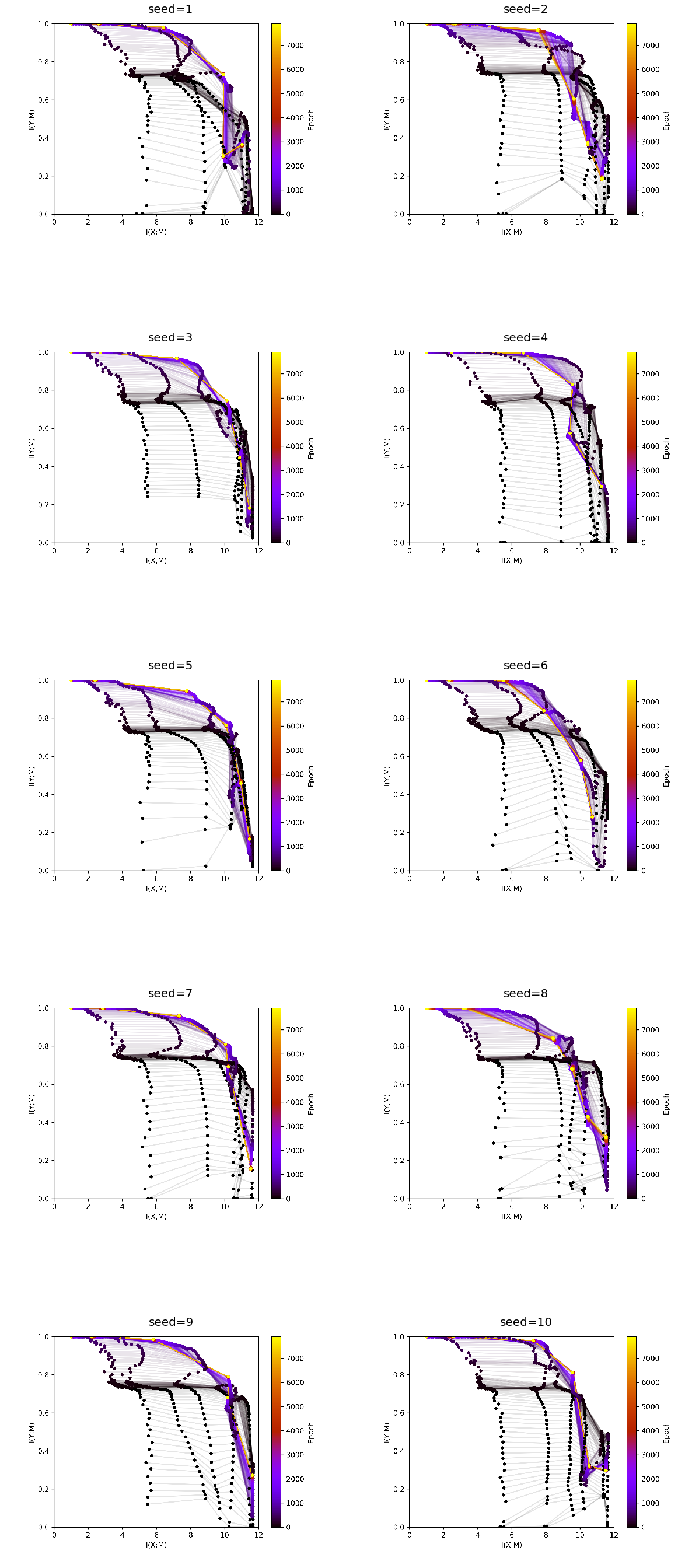
EDGE individiual runs¶
[17]:
fig, ax = plt.subplots(5,2, figsize = (20,50))
ax = ax.flat
experiment_ids = [1345,1344,1346,1352,1351,1348,1347,1353,1350,1349]
experiments = loader.find_by_ids(experiment_ids)
differing_config_keys = find_differing_config_keys(experiments)
for i, experiment in enumerate(experiments):
img = plt.imread(BytesIO(experiment.artifacts['infoplane_train'].content))
ax[i].axis('off')
ax[i].imshow(img)
ax[i].set_title(format_config(experiment.config, *differing_config_keys),
fontsize=20)
[18]:
plt.show()
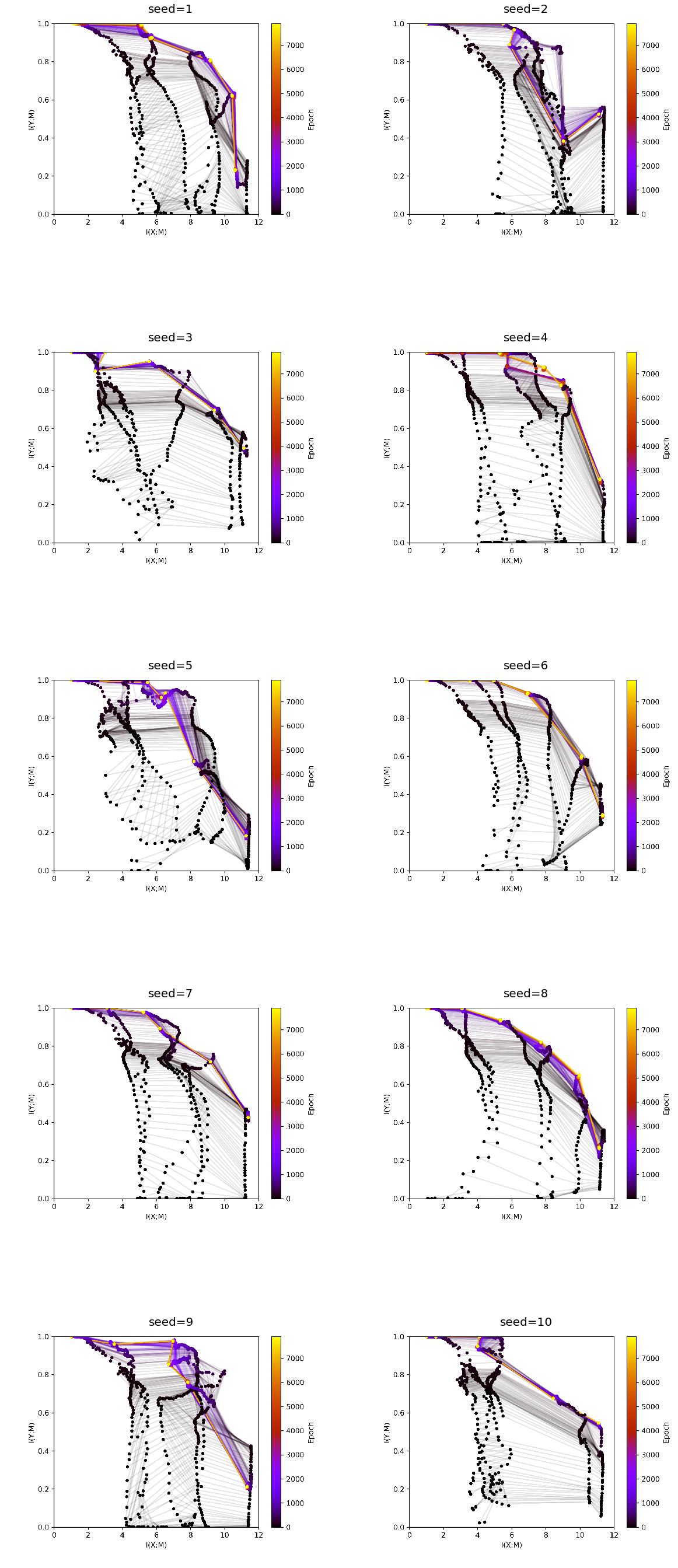
[19]:
fig, ax = plt.subplots(5,2, figsize = (20,50))
ax = ax.flat
experiment_ids = [1314,1339,1336,1338,1337,1335,1340,1341,1342,1343]
experiments = loader.find_by_ids(experiment_ids)
differing_config_keys = find_differing_config_keys(experiments)
for i, experiment in enumerate(experiments):
img = plt.imread(BytesIO(experiment.artifacts['infoplane_train'].content))
ax[i].axis('off')
ax[i].imshow(img)
ax[i].set_title(format_config(experiment.config, *differing_config_keys),
fontsize=20)
plt.show()
[ ]:
[ ]: